Data analysis II
Visualisation, practice
Laurent Bergé
University of Bordeaux, BxSE
09/12/2021
Outline
A tale of two paths...

Pros and cons: Base R
Pros
the principle is easy to grasp: we simply overlay successive forms on top of each other
low level operations = everything is possible
Pros and cons: Base R
Pros
the principle is easy to grasp: we simply overlay successive forms on top of each other
low level operations = everything is possible
Cons
because everything is an overlay on stg already in place, the first plot is critical: you need to plan everything in advance!!! Doing simple stuff can be surprisingly difficult.
many commands which are not really intuitive. After 10 years, I still look up
?parregularly.
Messing up the first plot in Base R
You miss most of the show!

Disclaimer
I'm a ggplot2 noob, base R is my home. I have a lot of sympathy for it.
Pros and cons: ggplot2
Pros
much more user friendly: stacks all your layers before creating the graph and makes all the computations for you. You don't need to overthink how to set the stage any more.
millions of contributed packages
Pros and cons: ggplot2
Pros
much more user friendly: stacks all your layers before creating the graph and makes all the computations for you. You don't need to overthink how to set the stage any more.
millions of contributed packages
Cons
- since there is pre-processing, it is sometimes difficult to do exactly what you want (e.g. for a very precise publication graph). To be confirmed, it's only second-hand experience!
Wait! There are other paths!

Ready made solutions
there are numerous packages out there which make good graphs with a user-friendly interface (ie with minimal user input)
example: ggpubr or fplot for distributions; ggcorrplot for correlations; highcharter for many things, etc.
Pros and cons: Ready made solutions
Pros
in a single line of code you get a graph of (usually) very decent quality
very good for exploratory graphs
Pros and cons: Ready made solutions
Pros
in a single line of code you get a graph of (usually) very decent quality
very good for exploratory graphs
Cons
- the level of customization is limited, this is especially problematic for presentations/publications in which you want a very high level of customization
Direct edition
I strongly encourage you to learn how to work with inkscape⋆
it's just crazy how fast you can edit/create images
that's indispensable in your skill set, you'll save so much time!
later: add gif edition of the castle image
Pros and cons: Direct edition
Pros
you can do exactly what you want, as precisely as you want
with practice, you can edit very rapidly
Pros and cons: Direct edition
Pros
you can do exactly what you want, as precisely as you want
with practice, you can edit very rapidly
Cons
the direct edition only comes after the first creation of the graph: hence you have to navigate across software
cannot be automated: all the work that you do with one graph, you'd have to do it again for a graph with new data
Short introduction to Base R
Data content
R dispose of multiple functions to display data:
plot(): CORE graphical functionpoints(),lines(),abline(),text(): used to plot additional datadensity(),hist(),boxplot(), etc...
Plot
The function
plot()is the main graphical function of R (more precisely, it's a method).By default it is a scatterplot between two variables, but it can be used to do much more than that.
Some functions preprocess the data, like
density(), and modify completely the behavior ofplot()when you apply it to the preprocessed data. More on that later.When you apply
plot(), it creates a new graphic and the previous one is lost (of course there are exceptions...). To add several pieces of information, you'll need to use other functions.
Main plot arguments
Main plot arguments relating to data:
x, y: the dataxlim, ylim: the limits of the plotting regioncol, pch, lty, lwd: color, symbol, line type and line widthtype: the type of plotlog: whether to put the x/y axes to logarithm
Plot: type
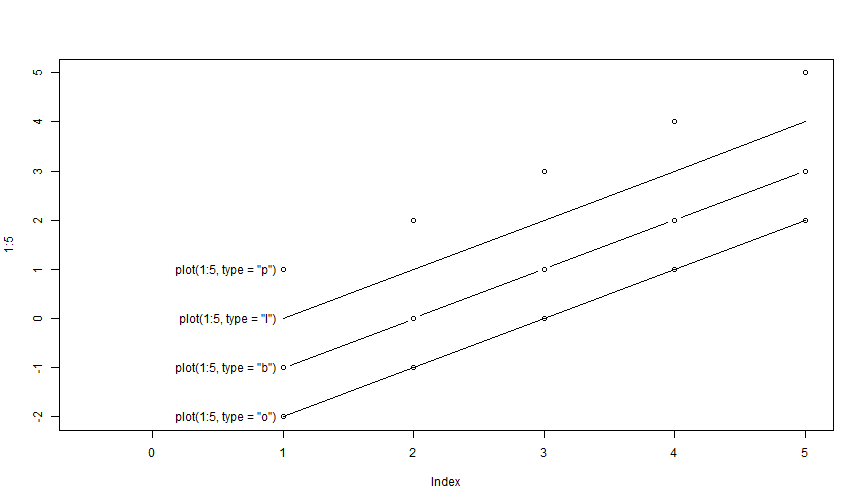
Plot: type = "n"
Using type = "n" hides the data, but EVERYTHING else is there. Can be useful when constructing complex graphics: i.e. when setting the stage.
plot(1:5, type = "n")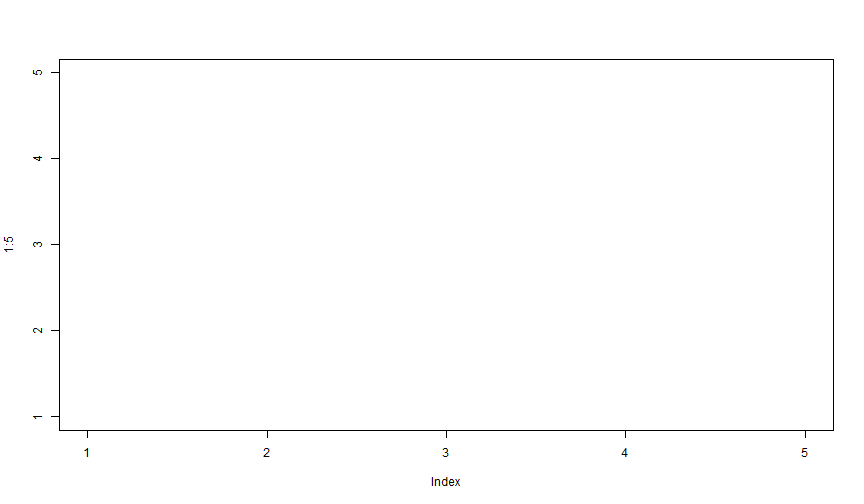
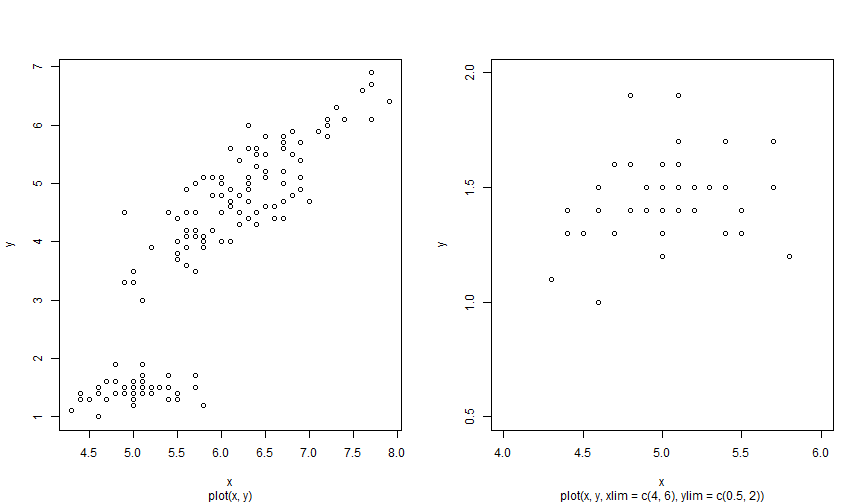
Plot: limits
Plot: pch
plot(1:20, pch = 1:20)grid()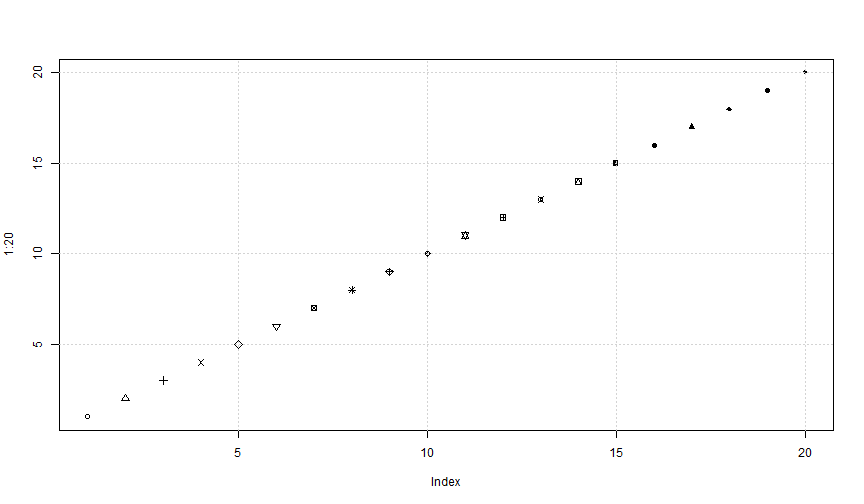
Plot: cex
plot(1:5, pch = 16, cex = 1:5, main = "cex: modify point size")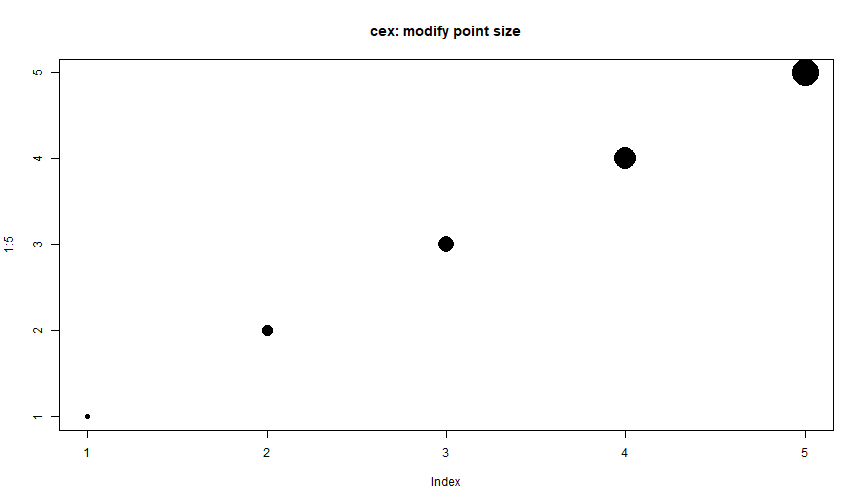
Plot: lty and lwd
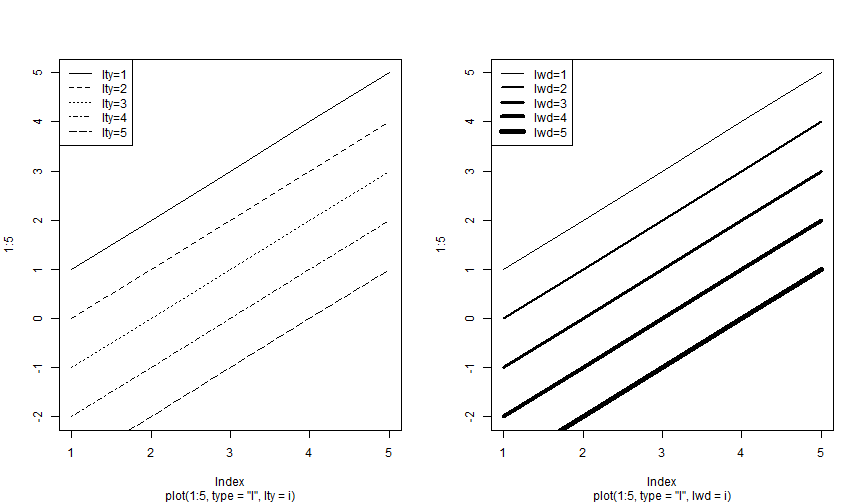
Plot: col I
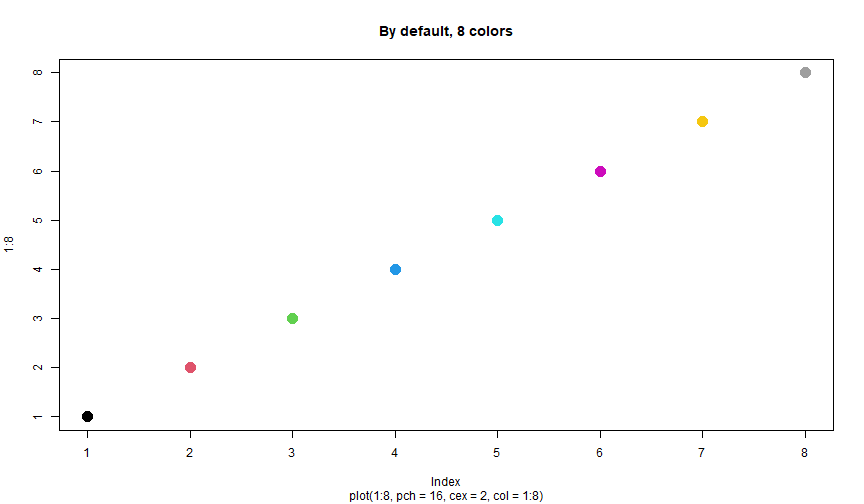
Plot: col II
Lot of color possibilities:
- Custom colors:
rgb(),hsv(), etc - "Nice" colors: package
RColorBrewer Color interpolation:
rainbow(n),heat.colors(n), etc, create vectors ofncolors.colorRampPalette(c("white", "blue"))(5): create a vector of 5 colors between the colors white and blue.
Nice introduction to R colors in the R-stats UBC course
Exercise: Plot
Generate a 100 periods Brownian motion xt+1=xt+ϵt, ϵt∼N(0,1).
- Plot its evolution with both a solid line and filled points (in the same graph).
- This time display only the points and use the function
rainbow()to set the color of each point.
Adding data points
To add points/lines onto an existing plot:
lines()points()
It behaves as the function plot() and contains the same arguments (col, lty, cex, lwd, pch).
Lines & points
plot(1:5, ylim = c(-2, 5))lines(1:5 - 1)points(1:5 - 2)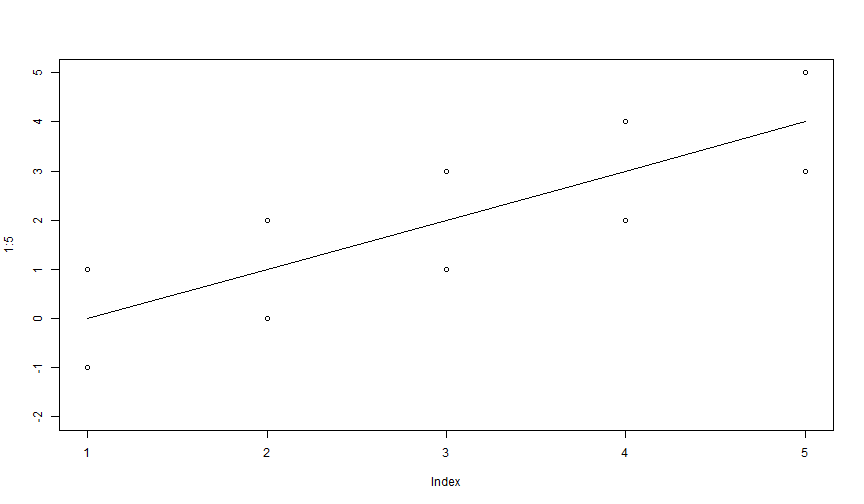
Exercise: Plot & line
In the following graph, the functions plot(), lines() and points() have been called. Can you say to what command refers each graphical information, and in what order they have been called?
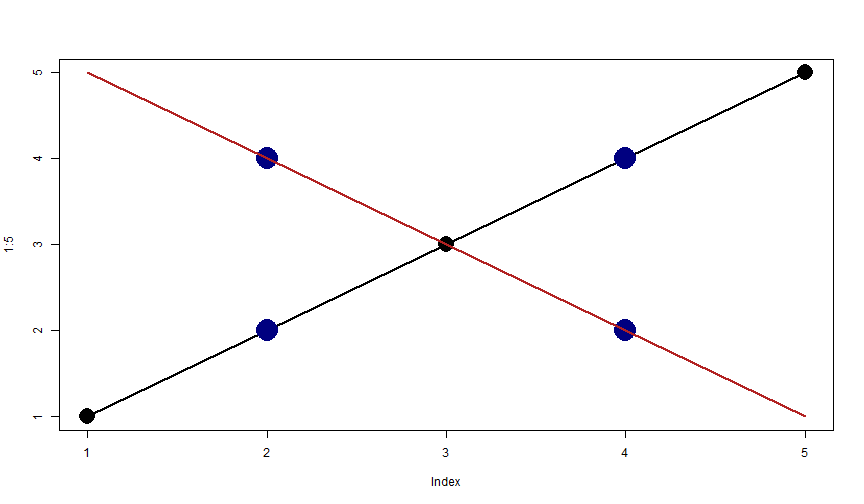
Exercise: Plot & lines
Re-generate the previous Brownian motion.
Plot it with both line and dots.
Generate another Brownian motion with ϵt∼N(0,4).
Plot the two motions on a single graph, the second one should be of"firebrick"color, have thick and dashed line and be of triangle symbol.
abline I
The function abline() draws lines. Its arguments are:
h: coordinate of horizontal linev: coordinate of vertical linea, b: intercept (a) and slope of a straight line. Shorthan exist: can take the result of an OLS regression (functionlm()) instead.
abline II
plot(iris$Sepal.Length, iris$Petal.Width)abline(lm(Petal.Width ~ Sepal.Length, iris))abline(h = c(1, 2), v = c(5, 7), col = "gray", lty = 3)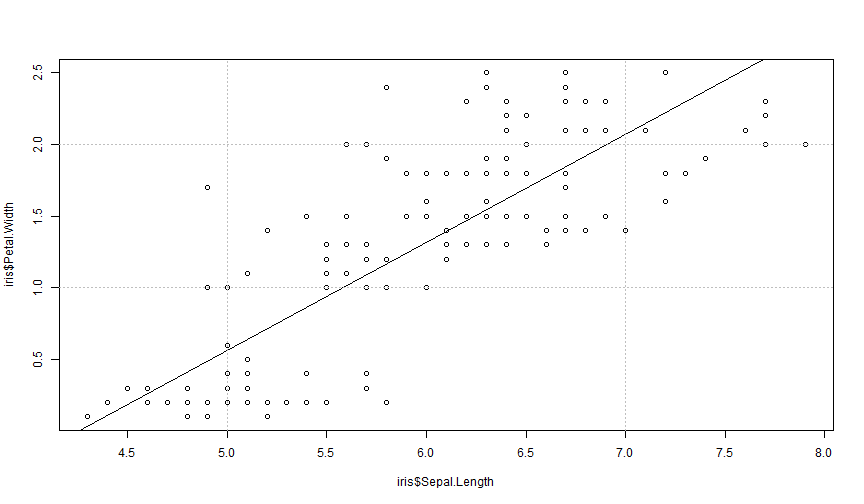
Exercise: abline
You want to illustrate the relation between the variables "Sepal.Length" and "Petal.Width" for each species of the iris data.
- Plot the scatterplot between the two variables with one color per species.
- Draw the regression lines for each group with the appropriate color.
Text I
You can add text to an existing plot with the function text(). The most important arguments are:
- x, y: coordinates of the text
- labels: the text to be displayed
- pos: the position of the text relative to the coordinate. pos = 0: as is, pos = 1: below, 2: left, 3: top, 4: right.
- As usual, other graphical parameters apply: cex (size), col, etc.
Text II
plot(5:1, col = "firebrick", pch = 18, xlim = c(0, 6))text(1, 5, "pos = default")text(2:5, 4:1, paste0("pos = ", 1:4), pos = 1:4)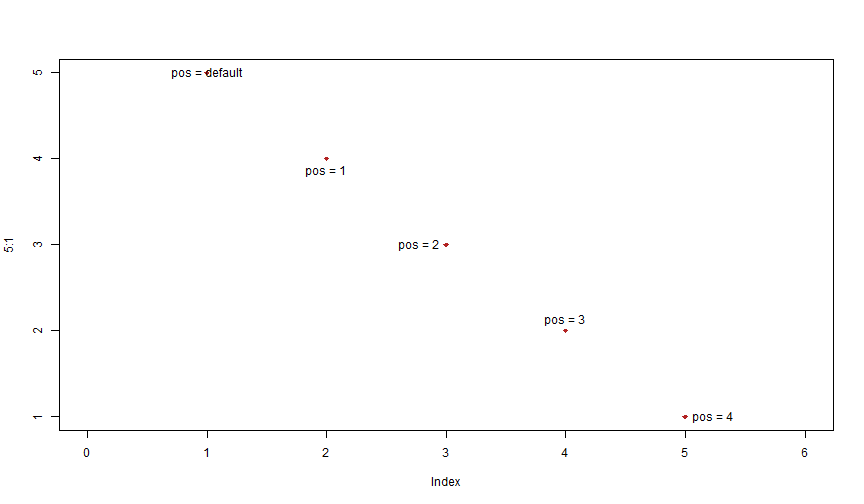
Exercise: text
- As in the previous exercise, plot the scatterplot between the two variables with one color per species for the variables "Sepal.Length" and "Petal.Width" of the
irisdata. - Add the Species names in the middle of the points for each species in the right color and with large font.
A functional approach to graphs
From pain we learn
Base R can be surprisingly painful for doing seemingly simple stuff.
From pain we learn
Base R can be surprisingly painful for doing seemingly simple stuff.
From pain we learn
Base R can be surprisingly painful for doing seemingly simple stuff.
Base R is so painful, that if you stick to it, it will make you a good programmer (or a masochist!).
Remember though: it's not just painful, it's also extremely powerful!
Hard coding
It's very easy to write code that is specific to your current data! In fact, it's usually the first thing we do, and it works well.
Hard coding
It's very easy to write code that is specific to your current data! In fact, it's usually the first thing we do, and it works well.
plot(iris$Petal.Length, iris$Sepal.Length, col = iris$Species, pch = 20, cex = 2)text(1.5, 5, "Setosa", font = 2, cex = 4)text(4, 6, "Versicolor", font = 2, cex = 4, col = 2)text(6, 7, "Virginica", font = 2, cex = 4, col = 3)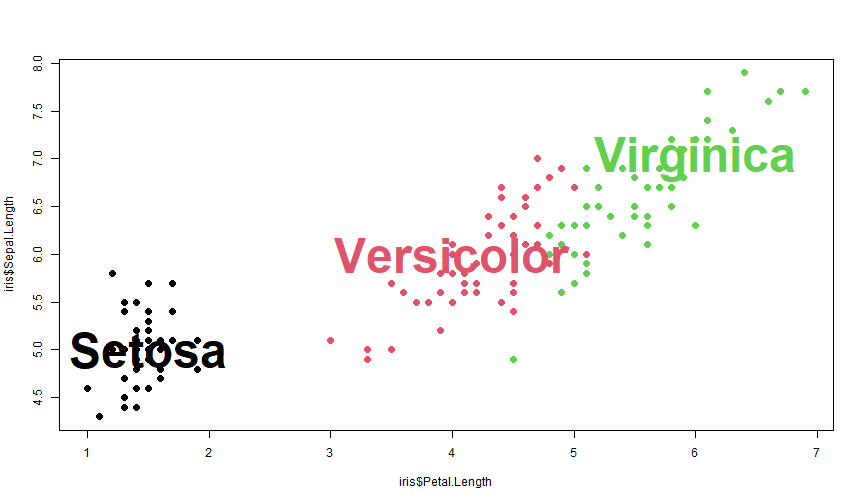
Hard coding: The problem
If your data changes, even slightly, your code is messed up.
Changing Sepal.Length into Sepal.Width loses the legend:
plot(iris$Petal.Length, iris$Sepal.Width, col = iris$Species, pch = 20, cex = 2)text(1.5, 5, "Setosa", font = 2, cex = 4)text(4, 6, "Versicolor", font = 2, cex = 4, col = 2)text(6, 7, "Virginica", font = 2, cex = 4, col = 3)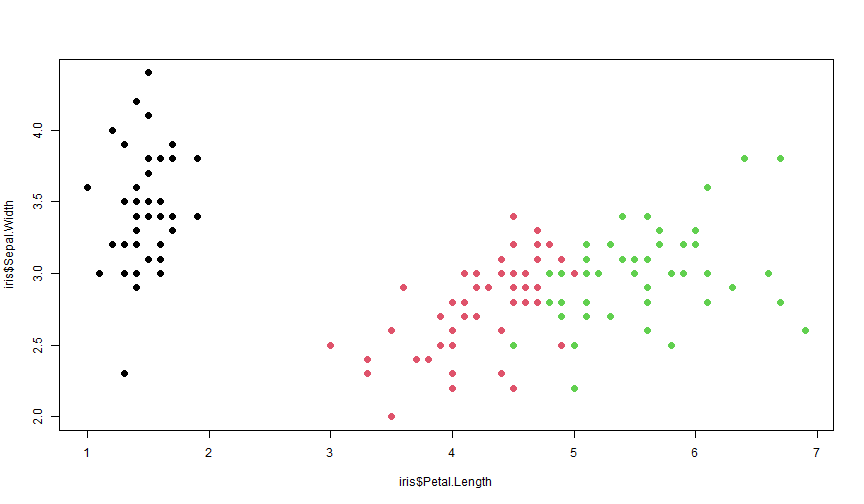
Hard coding: The problem
If your data changes, even slightly, your code is messed up.
The data always changes!
Hard coding: The routine
If you want to replicate a hard coded graph to a new data set you:
- copy paste the code
- change the data
- make the adjustments so the graph looks as you wish with the new data
Hard coding: The routine
If you want to replicate a hard coded graph to a new data set you:
- copy paste the code
- change the data
- make the adjustments so the graph looks as you wish with the new data
I think I don't need to write that each of these three steps are highly error-prone, and can cost dearly.⋆
Hard coding: The solution
- very simple: don't hard code!
Hard coding: The solution
very simple: don't hard code!
OK, here comes some tips
Tip 1: Define global variables!
- whenever a variable is repeated twice, use a global variable1 defined at the beginning of the piece of code
Tip 1: Define global variables!
- whenever a variable is repeated twice, use a global variable1 defined at the beginning of the piece of code
BAD
plot(iris$Petal.Length, iris$Sepal.Width, col = iris$Species, pch = 20, cex = 2)text(1.5, 5, "Setosa", font = 2, cex = 4)text(4, 6, "Versicolor", font = 2, cex = 4, col = 2)text(6, 7, "Virginica", font = 2, cex = 4, col = 3)GOOD
FONT = 2CEX = 4plot(iris$Petal.Length, iris$Sepal.Width, col = iris$Species, pch = 20, cex = 2)text(1.5, 5, "Setosa", font = FONT, cex = CEX)text(4, 6, "Versicolor", font = FONT, cex = CEX, col = 2)text(6, 7, "Virginica", font = FONT, cex = CEX, col = 3)Tip 2: Lay bare how you think!
when you decide to place some text here, or a legend there, how do you take the decision?
you decide based on heuristics (although you may not even notice there was a decision process!)
Tip 2: Lay bare how you think!
when you decide to place some text here, or a legend there, how do you take the decision?
you decide based on heuristics (although you may not even notice there was a decision process!)
the game is to extract the (often implicit) rules that made you take a decision⋆
if you achieve to make the heuristic explicit: you win since now you can automatize it!
Tip 2: Lay bare how you think!
Remember when I asked to put the names in the middle of the points?
Tip 2: Lay bare how you think!
Remember when I asked to put the names in the middle of the points?
Tip 2: Lay bare how you think!
Remember when I asked to put the names in the middle of the points?
BAD
FONT = 2CEX = 4plot(iris$Petal.Length, iris$Sepal.Width, col = iris$Species, pch = 20, cex = 2)text(1.5, 5, "Setosa", font = FONT, cex = CEX)text(4, 6, "Versicolor", font = FONT, cex = CEX, col = 2)text(6, 7, "Virginica", font = FONT, cex = CEX, col = 3)GOOD
FONT = 2CEX = 4plot(iris$Petal.Length, iris$Sepal.Width, col = iris$Species, pch = 20, cex = 2)bary = aggregate(cbind(Petal.Length, Sepal.Width) ~ Species, iris, mean)text(bary[1, 2], bary[1, 3], "Setosa", font = FONT, cex = CEX)text(bary[2, 2], bary[2, 3], "Versicolor", font = FONT, cex = CEX, col = 2)text(bary[3, 2], bary[3, 3], "Virginica", font = FONT, cex = CEX, col = 3)Tip 3: Loop whenever possible!
- whenever you repeat two statements: use a loop instead!
Tip 3: Loop whenever possible!
- whenever you repeat two statements: use a loop instead!
BAD
FONT = 2CEX = 4plot(iris$Petal.Length, iris$Sepal.Width, col = iris$Species, pch = 20, cex = 2)bary = aggregate(cbind(Petal.Length, Sepal.Width) ~ Species, iris, mean)text(bary[1, 2], bary[1, 3], "Setosa", font = FONT, cex = CEX)text(bary[2, 2], bary[2, 3], "Versicolor", font = FONT, cex = CEX, col = 2)text(bary[3, 2], bary[3, 3], "Virginica", font = FONT, cex = CEX, col = 3)GOOD
FONT = 2CEX = 4plot(iris$Petal.Length, iris$Sepal.Width, col = iris$Species, pch = 20, cex = 2)categ_val = levels(iris$Species)for(i in seq_along(categ_val)){ data = iris[iris$Species == categ_val[i], ] text(mean(data$Petal.Length), mean(data$Sepal.Width), categ_val[i], font = FONT, cex = CEX, col = i)}Tip 4: Loop over the tips!
Apply recursively Tip 1, Tip 2 and Tip 3 until you can't any more.
Tip 4: Loop over the tips!
Apply recursively Tip 1, Tip 2 and Tip 3 until you can't any more.
BAD
FONT = 2CEX = 4plot(iris$Petal.Length, iris$Sepal.Width, col = iris$Species, pch = 20, cex = 2)categ = levels(iris$Species)for(i in seq_along(categ)){ data = iris[iris$Species == categ[i], ] text(mean(data$Petal.Length), mean(data$Sepal.Width), categ[i], font = FONT, cex = CEX, col = i)}GOOD
FONT = 2CEX = 4x = iris$Petal.Lengthy = iris$Sepal.Widthcateg = iris$Speciesplot(x, y, col = categ, pch = 20, cex = 2)categ_val = levels(categ)for(i in seq_along(categ_val)){ who = categ == categ_val[i] text(mean(x[who]), mean(y[who]), categ_val[i], font = FONT, cex = CEX, col = i)}Are the tips useful?
Can those tips be concretely helpful?
To know that, let's summon the copy-paste demon.
Code without tips
plot(iris$Petal.Length, iris$Sepal.Width, col = iris$Species, pch = 20, cex = 2)text(1.5, 5, "Setosa", font = 2, cex = 4)text(4, 6, "Versicolor", font = 2, cex = 4, col = 2)text(6, 7, "Virginica", font = 2, cex = 4, col = 3)Code without tips: Summoning

Code without tips: Outcome
The demon has immense powers

Code with tips
FONT = 2CEX = 4x = iris$Petal.Lengthy = iris$Sepal.Widthcateg = iris$Speciesplot(x, y, col = categ, pch = 20, cex = 2)categ_val = levels(categ)for(i in seq_along(categ_val)){ who = categ == categ_val[i] text(mean(x[who]), mean(y[who]), categ_val[i], font = FONT, cex = CEX, col = i)}Code with tips: Summoning

Code with tips: Outcome
The demon is weak


Tips: Side benefits
If you've followed the tips, guess what:
Tips: Side benefits
If you've followed the tips, guess what:
You can create a function for your graph for free!
Tips: Side benefits
If you've followed the tips, guess what:
You can create a function for your graph for free!
Before
FONT = 2CEX = 4x = iris$Petal.Lengthy = iris$Sepal.Widthcateg = iris$Speciesplot(x, y, col = categ, pch = 20, cex = 2)categ_val = levels(categ)for(i in seq_along(categ_val)){ who = categ == categ_val[i] text(mean(x[who]), mean(y[who]), categ_val[i], font = FONT, cex = CEX, col = i)}After
scatter_name = function(x, y, categ, font = 2, cex = 4){ plot(x, y, col = categ, pch = 20, cex = 2) categ_val = levels(categ) for(i in seq_along(categ_val)){ who = categ == categ_val[i] text(mean(x[who]), mean(y[who]), categ_val[i], font = font, cex = cex, col = i) }}scatter_name(iris$Petal.Length, iris$Sepal.Width, iris$Species)Why create functions to make graphs?
- guards you against, or limits, copy-paste problems
Why create functions to make graphs?
guards you against, or limits, copy-paste problems
facilitates graph replications
Why create functions to make graphs?
guards you against, or limits, copy-paste problems
facilitates graph replications
you don't have to think to implementation details when running the function (reduces mental load)
Why create functions to make graphs?
guards you against, or limits, copy-paste problems
facilitates graph replications
you don't have to think to implementation details when running the function (reduces mental load)
it's very easy to include new features to the functions, and all the calls benefit from it
Mental load
Code telling what you do and not how you do it increases productivity tremendously.
Mental load
Code telling what you do and not how you do it increases productivity tremendously.
FONT = 2CEX = 4x = iris$Petal.Lengthy = iris$Sepal.Widthcateg = iris$Speciesplot(x, y, col = categ, pch = 20, cex = 2)categ_val = levels(categ)for(i in seq_along(categ_val)){ who = categ == categ_val[i] text(mean(x[who]), mean(y[who]), categ_val[i], col = i, font = FONT, cex = CEX)}scatter_name(iris$Petal.Length, iris$Sepal.Width, iris$Species)Mental load
Code telling what you do and not how you do it increases productivity tremendously.
FONT = 2CEX = 4x = iris$Petal.Lengthy = iris$Sepal.Widthcateg = iris$Speciesplot(x, y, col = categ, pch = 20, cex = 2)categ_val = levels(categ)for(i in seq_along(categ_val)){ who = categ == categ_val[i] text(mean(x[who]), mean(y[who]), categ_val[i], col = i, font = FONT, cex = CEX)}scatter_name(iris$Petal.Length, iris$Sepal.Width, iris$Species)The code on the right will always be easier to understand than the code on the left.⋆
Why not create functions?
- you have a presentation in 30 minutes and have to finish that graph
Why not create functions?
you have a presentation in 30 minutes and have to finish that graph
you're making a graph that you think will never replicate⋆
Why not create functions?
you have a presentation in 30 minutes and have to finish that graph
you're making a graph that you think will never replicate⋆
the graph is really simple (in terms of lines of code!)
Functions: Summary
thinking in functions will change the way you code
it will clarify your code: it will be easier to understand and share, and less error-prone
due to the high fixed costs, 0 marginal cost nature of functions, you'll gain a lot of productivity
Functional programming: Application
Remember the scatterplot with different colors and a linear fit? Let's redo it.
- plot the scatterplot between the variables "Sepal.Length" and "Petal.Width" for each species of the
irisdata, and add a linear fit
Functional programming: Application
Remember the scatterplot with different colors and a linear fit? Let's redo it.
plot the scatterplot between the variables "Sepal.Length" and "Petal.Width" for each species of the
irisdata, and add a linear fituse
segments()to shorten the fit to the width of the scatterplot
Functional programming: Application
Remember the scatterplot with different colors and a linear fit? Let's redo it.
plot the scatterplot between the variables "Sepal.Length" and "Petal.Width" for each species of the
irisdata, and add a linear fituse
segments()to shorten the fit to the width of the scatterplottransform it into a function, with the appropriate arguments
Functional programming: Application
Remember the scatterplot with different colors and a linear fit? Let's redo it.
plot the scatterplot between the variables "Sepal.Length" and "Petal.Width" for each species of the
irisdata, and add a linear fituse
segments()to shorten the fit to the width of the scatterplottransform it into a function, with the appropriate arguments
add the argument
line_extendgiving how much the length of the segment should be extended, in % of the graph width (default is 0)
Functional programming: Application
Remember the scatterplot with different colors and a linear fit? Let's redo it.
plot the scatterplot between the variables "Sepal.Length" and "Petal.Width" for each species of the
irisdata, and add a linear fituse
segments()to shorten the fit to the width of the scatterplottransform it into a function, with the appropriate arguments
add the argument
line_extendgiving how much the length of the segment should be extended, in % of the graph width (default is 0)control the arguments given by the user
More Base R graphs stuff
Informative content
So far we've seen only data content. But there's much more to make a good graph: all the surrounding information!
plot()arguments:xlab,ylab: x/y axis labelssub,main: subtitle and main titleaxes: whether to draw the axesann: ifFALSE, cleans all x/y labels
legend(): adds a legendtitle(): adds axis labels and titles (close to previous plot arguments)axis(): function to draw the axes of a plot.- mathematical formulas
Plot: informative
plot(-1:1, -1:1, xlab = "xlab", ylab = "ylab", main = "main", sub = "sub", type = "n")text(0, 0, 'plot(-1:1, -1:1, xlab = "xlab", ylab = "ylab", main = "main", sub = "sub")')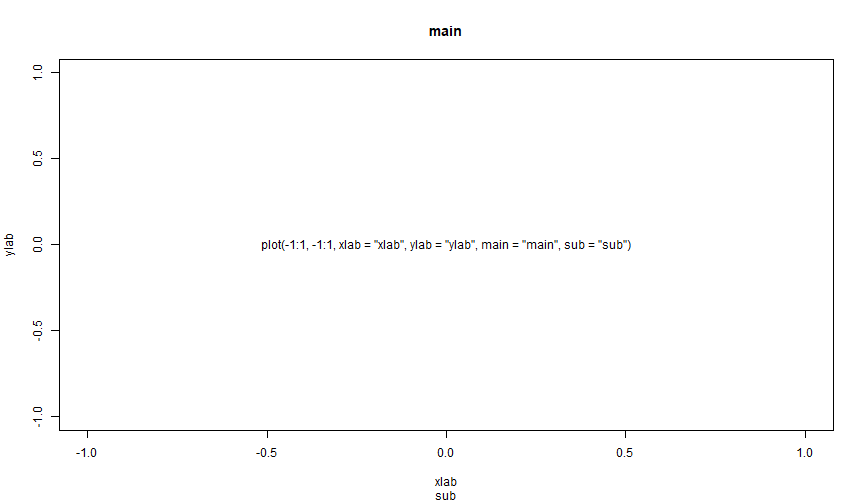
Exercise: informative content
Do the scatterplot between variables "Sepal.Length" and "Petal.Width" of the iris data.
- Put appropriate axes labels (i.e. add only the name of the variable).
- Put the correlation in the title of the plot.
Title
You can add a title after a plot is done with title().
plot(1:5)title(main = "This is the title", sub = "This is the subtitle")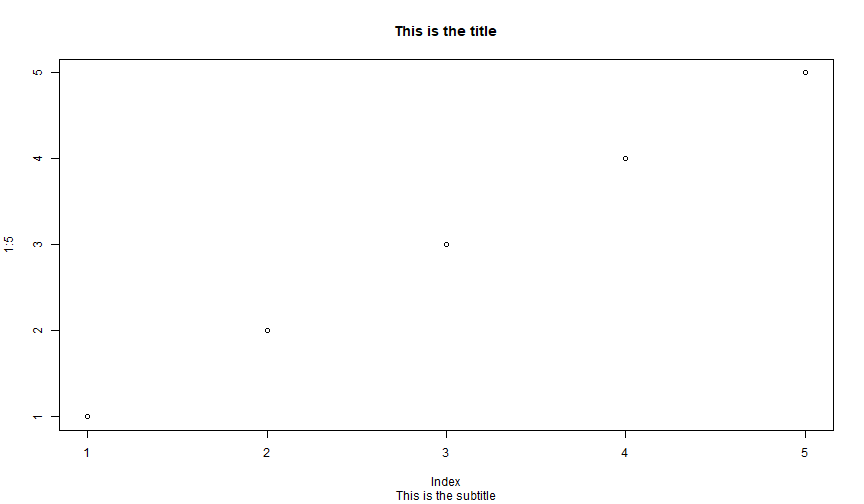
Title: mtext
Use mtext to add text in the margin of the graph. Can be used to insert a title.
plot(1:5)mtext("That's a basic graph", side = 3, line = 1, font = 2, adj = 0)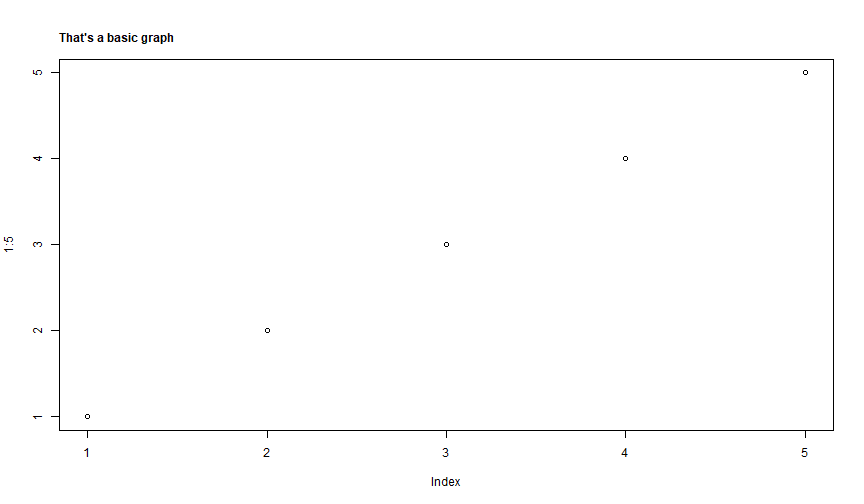
Adding a legend
You can add a legend to clarify the content of a plot. A legend is a piece of information appearing inside the plotting region.
Here are the main arguments:
x,y: the location of the legend (top left corner). There exist shorthands! instead you can use "topleft", "right", etc.legend: the content of the legend (a character vector.)pch,lty,col,lwd: thepch,lty,col,lwdassociated to the legend vectorbty: whether or not to show the legend box ("o" is default, "n" removes it)
Keep in mind that there are many more arguments.
Legend
Legend in the bottom I
How to have a legend in the bottom? Here's some ready-made code.
legend_bottom = function(..., bty = "n"){ # Original credits to: https://stackoverflow.com/questions/3932038/plot-a-legend-outside-of-the-plotting-area-in-base-graphics/3932558 op = par(fig = c(0, 1, 0, 1), oma = c(0, 0, 0, 0), mar = c(0, 0, 0, 0), new = TRUE) on.exit(par(op)) plot(0, 0, type = 'n', axes = FALSE, ann = FALSE) legend("bottom", horiz = TRUE, bty = bty, ...)}Legend in the bottom II
plot(iris$Sepal.Length, iris$Petal.Width, col = iris$Species, pch = 15)legend_bottom(legend = levels(iris$Species), col = 1:3, pch = 15)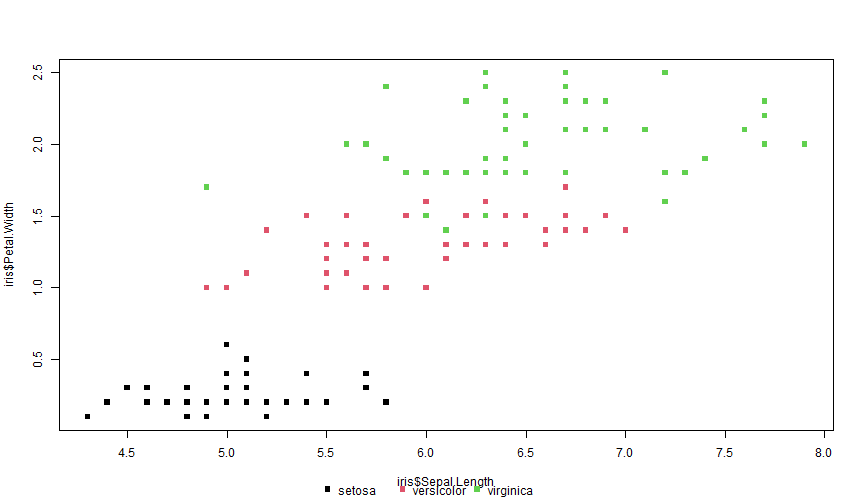
Legend in the bottom II
plot(iris$Sepal.Length, iris$Petal.Width, col = iris$Species, pch = 15)legend_bottom(legend = levels(iris$Species), col = 1:3, pch = 15)
Oh, yeah the legend ends up being too close to the label... Remember about "setting the stage"? To make it nicer, you'd need to increase the bottom margin beforehand with, e.g., par(mar = c(7, 4, 2, 2)) :-/ That's one of the reasons why ggplot is so much easier to handle.
Axes
You can modify the axes at will.
axis(i) draws the ith axis with, 1: bottom, 2: left, 3: top and 4: right.
plot(1:5, axes = FALSE)axis(1)axis(4)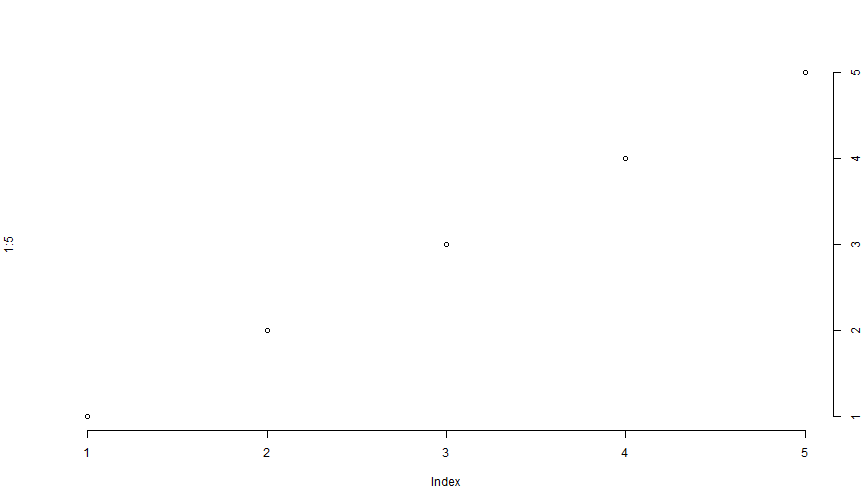
Axis
The function axis has the following main options:
side: where to draw the axis (1: bottom, etc, 4:right)at: where the ticks are drawnlabels: the labels at the ticks (usually numbers)lwd: line width of the horizontal line (if side is 1 or 3)lwd.ticks: the line width of the ticks. Default is equal to lwdtck: length of the ticks (in fraction of the plotting region)las: orientation of the text
Many other options.
Axis example
plot(1:5, axes = FALSE, ann = FALSE)box() # draw a simple boxaxis(1, 1:4, c("First", "Second", "Third", "Fourth"), cex.axis = .9)axis(3, 5, "Fifth", lwd.ticks = 2)axis(2, 1:3, c("One", "Two", "Three"), las = 2)axis(2, 4, "Four", col.axis = "red")axis(2, 5, "Five", col.ticks = "blue", lwd = 2)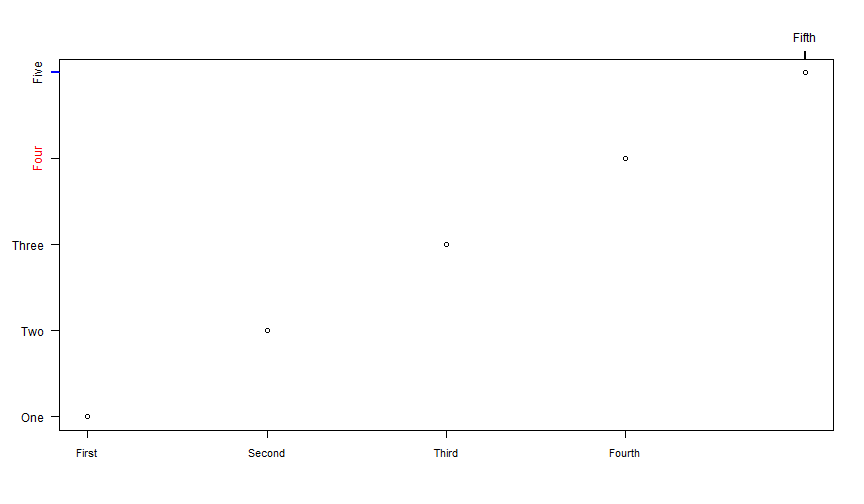
Mathematical expressions
You can add mathematical expressions in graphics. Advice: for single mathematical formulas, use only function substitute().
Write a formula inside the function substitute() (a bit like in Latex). Some elements that can compose the formula:
x[i]for subscript xix^ifor superscript xix %in% y, x∈yalpha,beta,gamma, etc...paste(x, y, z): juxtapose the three components
See ?plotmath for more details.
Math
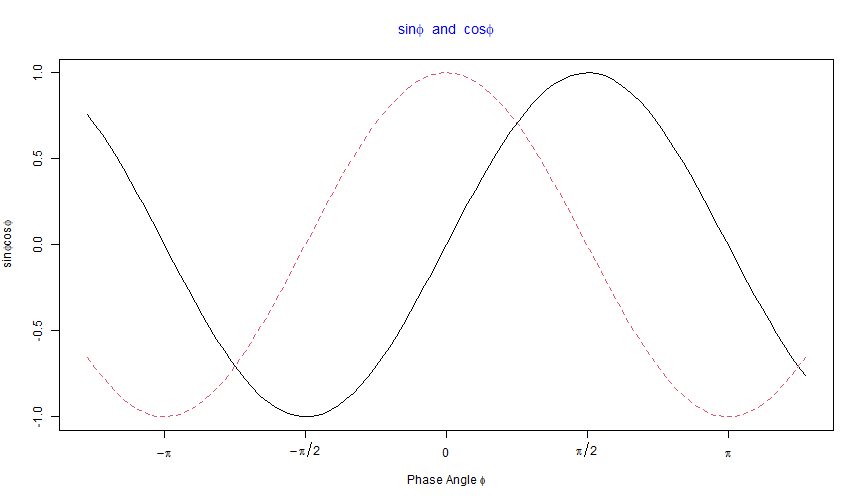
substitute
substitute() contains a second argument. It can be used to replace some variables with numbers:
curve(sin(x)*sqrt(x), 1, 10000, log = "x", axes = FALSE, ann = FALSE)title(main = substitute(sin(x)%*%sqrt(x)))box() ; axis(2)for(i in 0:4) axis(1, at = 10**i, substitute(10^p, list(p = i)), cex.axis = 2)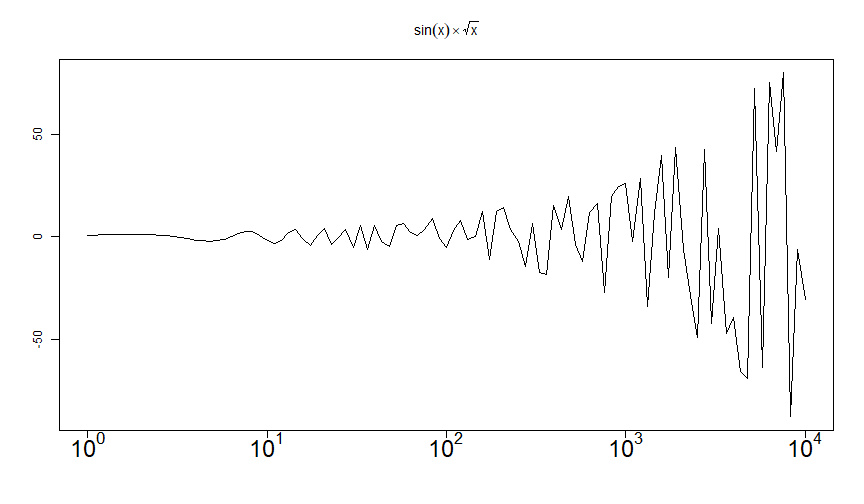
Graphical parameters
The function par() contains most graphical parameters (there are 72... ).
You can change the graphical parameters directly with par().
par(cex = 2, lwd = 2)plot(1:5, type = "o")
par
All plots will have these parameters as default. To reinitialize it, you can:
- reinitialize it manually:
par(cex = 1, lwd = 1) - Make a "save" when you modify it:
op = par(cex = 2, lwd = 2) # save old params# make the graphspar(op) # reinitialize itpar: other parameters
Some useful parameters:
mar: the margins of the plot: the space between the axes and the edge of the plot. It's a vector of length 4 (1st is bottom, last is right).cex,cex.axis,cex.lab,cex.main,cex.sub: expansion factor for different situationscol,col.axis,col.lab,col.main,col.sub.bg: color of the background (default is white)family: font family: can be "serif", "sans" and "mono"las: orientation of text (1: horizontal, 3:vertical)
See ?par for more details.
Multiple graphs
You can combine multiple graphs in one. Simplest way is to use mfrow:
op = par(mfrow = c(2, 2))for(i in 1:4) curve(sin(x) * x**i, -10, 10, ylab = paste0("sin(x) * x^", i))par(op) # reset to an unique frame
Conclusion
I'm afraid you won't be able to make nice graphs from this bare bones introduction!
It only brushes the topic but I hope that you could get some insights along the way! And especially that the functional programming approach convinced you!
Cheers!